Class for computing a feature transform used for preconditioning of the training data in neural-networks. More...
#include <get-feature-transform.h>
Public Member Functions | |
| void | Estimate (const FeatureTransformEstimateOptions &opts, Matrix< BaseFloat > *M, TpMatrix< BaseFloat > *within_cholesky) const |
| Estimates the LDA transform matrix m. More... | |
 Public Member Functions inherited from LdaEstimate Public Member Functions inherited from LdaEstimate | |
| LdaEstimate () | |
| void | Init (int32 num_classes, int32 dimension) |
| Allocates memory for accumulators. More... | |
| int32 | NumClasses () const |
| Returns the number of classes. More... | |
| int32 | Dim () const |
| Returns the dimensionality of the feature vectors. More... | |
| void | ZeroAccumulators () |
| Sets all accumulators to zero. More... | |
| void | Scale (BaseFloat f) |
| Scales all accumulators. More... | |
| double | TotCount () |
| Return total count of the data. More... | |
| void | Accumulate (const VectorBase< BaseFloat > &data, int32 class_id, BaseFloat weight=1.0) |
| Accumulates data. More... | |
| void | Estimate (const LdaEstimateOptions &opts, Matrix< BaseFloat > *M, Matrix< BaseFloat > *Mfull=NULL) const |
| Estimates the LDA transform matrix m. More... | |
| void | Read (std::istream &in_stream, bool binary, bool add) |
| void | Write (std::ostream &out_stream, bool binary) const |
Static Protected Member Functions | |
| static void | EstimateInternal (const FeatureTransformEstimateOptions &opts, const SpMatrix< double > &total_covar, const SpMatrix< double > &between_covar, const Vector< double > &mean, Matrix< BaseFloat > *M, TpMatrix< BaseFloat > *C) |
 Static Protected Member Functions inherited from LdaEstimate Static Protected Member Functions inherited from LdaEstimate | |
| static void | AddMeanOffset (const VectorBase< double > &total_mean, Matrix< BaseFloat > *projection) |
| This function modifies the LDA matrix so that it also subtracts the mean feature value. More... | |
Additional Inherited Members | |
 Protected Member Functions inherited from LdaEstimate Protected Member Functions inherited from LdaEstimate | |
| void | GetStats (SpMatrix< double > *total_covar, SpMatrix< double > *between_covar, Vector< double > *total_mean, double *sum) const |
| Extract a more processed form of the stats. More... | |
| LdaEstimate & | operator= (const LdaEstimate &other) |
 Protected Attributes inherited from LdaEstimate Protected Attributes inherited from LdaEstimate | |
| Vector< double > | zero_acc_ |
| Matrix< double > | first_acc_ |
| SpMatrix< double > | total_second_acc_ |
Class for computing a feature transform used for preconditioning of the training data in neural-networks.
By preconditioning here, all we really mean is an affine transform of the input data– say if we set up the classification as going from vectors x_i to labels y_i, then this would be a linear transform on X, so we replace x_i with x'_i = A x_i + b. The statistics we use to obtain this transform are the within-class and between class variance statistics, and the global data mean, that we would use to estimate LDA. When designing this, we had a few principles in mind:
Basically our method is as follows:
We need to explain the step that applies the dimension-specific scaling, which we described above as, "Apply a transform that reduces the variance of dimensions with low between-class variance". For a particular dimension, let the between-class diagonal covariance element be , and the within-class diagonal covariance is 1 at this point (since we have normalized the within-class covariance to unity); hence, the total variance is + 1. Below, "within-class-factor" is a constant that we set by default to 0.001. We scale the i'th dimension of the features by:
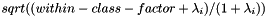
If >> 1, this scaling factor approaches 1 (we don't need to scale up dimensions with high between-class variance as they already naturally have a higher variance than other dimensions. As becomes small, this scaling factor approaches sqrt(within-class-factor), so dimensions with very small between-class variance get assigned a small variance equal to within-class-factor, and for dimensions with intermediate between-class variance, they end up with a variance roughly equal to : consider that the variance was originally (1 + ), so by scaling the features by approximately sqrt(() / (1 + )), the variance becomes approximately [this is clear after noting that the variance gets scaled by the square of the feature scale].
Definition at line 133 of file get-feature-transform.h.
| void Estimate | ( | const FeatureTransformEstimateOptions & | opts, |
| Matrix< BaseFloat > * | M, | ||
| TpMatrix< BaseFloat > * | within_cholesky | ||
| ) | const |
Estimates the LDA transform matrix m.
If Mfull != NULL, it also outputs the full matrix (without dimensionality reduction), which is useful for some purposes. If opts.remove_offset == true, it will output both matrices with an extra column which corresponds to mean-offset removal (the matrix should be multiplied by the feature with a 1 appended to give the correct result, as with other Kaldi transforms.) "within_cholesky" is a pointer to an SpMatrix that, if non-NULL, will be set to the Cholesky factor of the within-class covariance matrix. This is used for perturbing features.
Definition at line 28 of file get-feature-transform.cc.
References count, FeatureTransformEstimate::EstimateInternal(), LdaEstimate::GetStats(), and KALDI_LOG.
Referenced by main().
|
staticprotected |
Definition at line 40 of file get-feature-transform.cc.
References SpMatrix< Real >::AddMat2Sp(), MatrixBase< Real >::AddMatMat(), LdaEstimate::AddMeanOffset(), SpMatrix< Real >::AddSp(), VectorBase< Real >::ApplyCeiling(), TpMatrix< Real >::Cholesky(), MatrixBase< Real >::CopyFromMat(), TpMatrix< Real >::CopyFromTp(), FeatureTransformEstimateOptions::dim, VectorBase< Real >::Dim(), rnnlm::i, MatrixBase< Real >::Invert(), KALDI_ASSERT, KALDI_LOG, kaldi::kNoTrans, kaldi::kTrans, VectorBase< Real >::Max(), FeatureTransformEstimateOptions::max_singular_value, MatrixBase< Real >::MulRowsVec(), rnnlm::n, MatrixBase< Real >::NumCols(), MatrixBase< Real >::NumRows(), PackedMatrix< Real >::NumRows(), MatrixBase< Real >::Range(), FeatureTransformEstimateOptions::remove_offset, TpMatrix< Real >::Resize(), Matrix< Real >::Resize(), MatrixBase< Real >::Row(), kaldi::SortSvd(), MatrixBase< Real >::Svd(), SpMatrix< Real >::Trace(), and FeatureTransformEstimateOptions::within_class_factor.
Referenced by FeatureTransformEstimate::Estimate(), and FeatureTransformEstimateMulti::EstimateTransformPart().